
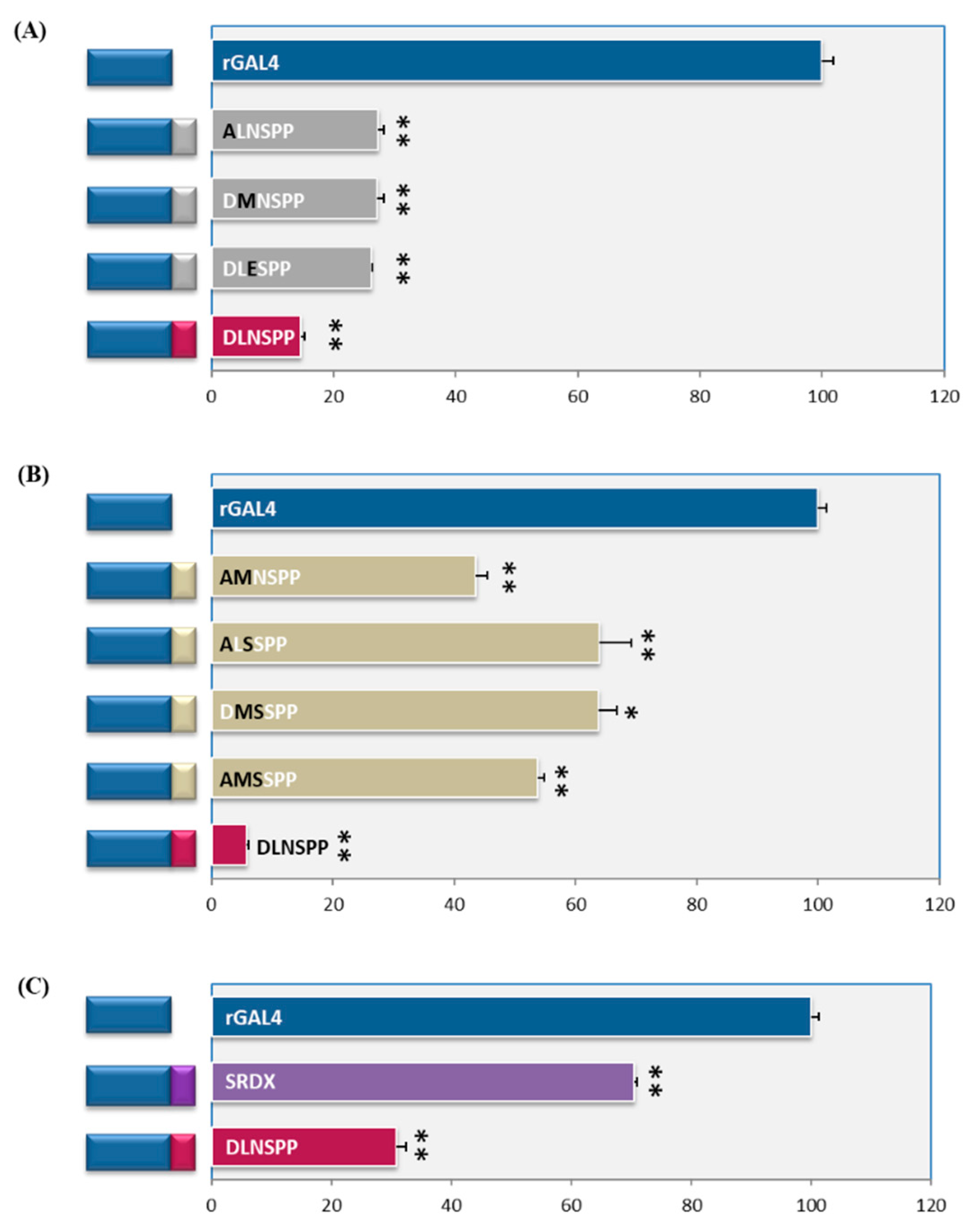
The OsPIL11-SRDX and OsPIL16-SRDX seedlings grown in darkness had constitutively photomorphogenic phenotypes with short coleoptiles and open leaf blades. In the present study, to explore the functions of PILs in rice skotomorphogenesis, we generated OsPIL11-SRDX and OsPIL16-SRDX transgenic lines by fusing the SRDX transcriptional repressor motif to the C-terminal of two members of the phytochrome interacting factor-like (OsPIL) family in rice (OsPIL11 and OsPIL16).
#Srdx recognition domain Activator#
It has been reported that SRDX motif, LDLDLELRLGFA, was able to convert a transcriptional activator into a strong repressor. Because of the development of rice direct seeding cultivation systems, there is an increasing need for clarifying the molecular mechanism underlying rice skotomorphogenic development. The hierarchal model described here for Vaccinium may also occur in a wider group of plants as a means to co-regulate different branches of the flavonoid pathway.ĭark-grown seedlings develop skotomorphogenesis. In contrast, VmMYBA1 is well expressed in bilberry flesh, up-regulating VmMYBPA1.1, allowing coordinated regulation of flavonoid biosynthesis genes and anthocyanin accumulation. Our findings suggest that the lack of VcMYBA1 expression in blueberry flesh results in an absence of VcMYBPA1.1 expression, which are both required for anthocyanin regulation. The MYBPA1.1 promoter is directly activated by MYBA1 and MYBPA2 proteins, which regulate anthocyanins and proanthocyanidins, respectively. MYBPA1.1 was unable to induce anthocyanin or proanthocyanidin accumulation in Nicotiana benthamiana, but activated promoters of flavonoid biosynthesis genes. MYBPA1.1 had a biphasic expression profile, correlating with both proanthocyanidin biosynthesis early during fruit development and anthocyanin biosynthesis during berry ripening. Comparative transcriptomics between blueberry and bilberry revealed that MYBPA1.1 and MYBA1 strongly correlated with the presence of anthocyanins, but were absent or weakly expressed in blueberry flesh. corymbosum) has white flesh, while bilberry (V. Members of the Vaccinium genus bear fruits rich in anthocyanins, a class of red-purple flavonoid pigments that provide human health benefits, although the localization and concentrations of anthocyanins differ between species: blueberry (V. Thus, it should be useful not only for the rapid analysis of the functions of redundant plant transcription factors but also for the manipulation of plant traits via the suppression of gene expression that is regulated by specific transcription factors. This chimeric repressor silencing technology (CRES-T), exploiting the EAR-motif repression domain, is simple and effective and can overcome genetic redundancy. Chimeric EIN3, CUC1, PAP1, and AtMYB23 repressors that included the EAR motif dominantly suppressed the expression of their target genes and caused insensitivity to ethylene, cup-shaped cotyledons, reduction in the accumulation of anthocyanin, and absence of trichomes, respectively. We show here that four different transcription factors fused to the EAR motif, a repression domain of only 12 amino acids, act as dominant repressors in transgenic Arabidopsis and suppress the expression of specific target genes, even in the presence of the redundant transcription factors, with resultant dominant loss-of-function phenotypes. Extensive experiments are conducted on the DREAMER and DEAP datasets, and the results show the superiority of our proposed method.The redundancy of genes for plant transcription factors often interferes with efforts to identify the biologic functions of such factors. We compute the SPD matrix based on the covariance as a feature and make a novel attempt to combine prototype learning with the Riemannian metric. Our method jointly exploits feature adaptation with distribution confusion and sample adaptation with centroid alignment. To solve this problem, we propose a domain adaptation SPD matrix network (daSPDnet) that can successfully capture an intrinsic emotional representation shared between different subjects. Recently, developing an effective brain-computer interface with a short calibration time has become a challenge. The conventional method involves the collection of a large amount of calibration data to build subject-specific models. Due to individual variability, training a generic emotion recognition model across different subjects is difficult. Emotion plays a vital role in human daily life, and EEG signals are widely used in emotion recognition.


 0 kommentar(er)
0 kommentar(er)
